Chemical Descriptors
Similar to chemists, models provide better predictions if they can understand similarities and differences between chemical compounds. The best way to to give models chemical intuition is using chemical descriptors.
Chemical descriptors are vectors (collections of numbers) describing properties of individual compounds. For example, relevant properties of solvents include polarity, or the ability to donate and receive hydrogen bonds.
Ultimately, descriptors let the model navigate the reaction space in a systematic and informed way.
Generating Descriptors from SMILES
The simplest way to generate descriptors is using the Generate Descriptors button on the Reaction Parameters page.
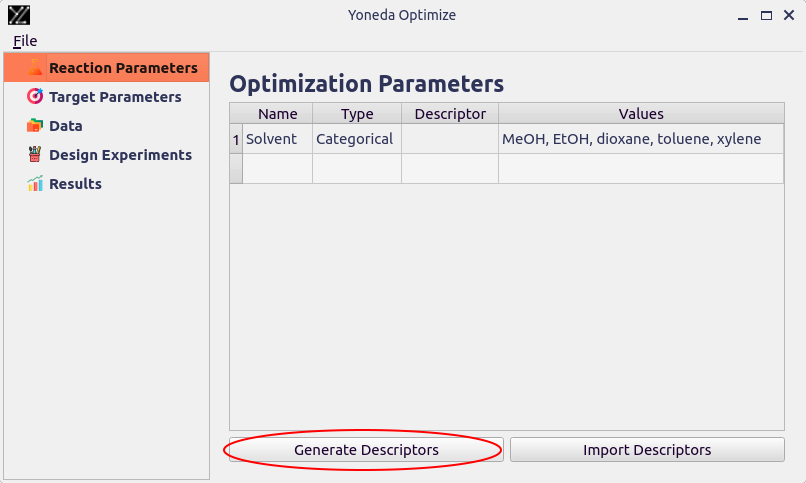
You will be prompted to select which parameter you would like to generate descriptors for.
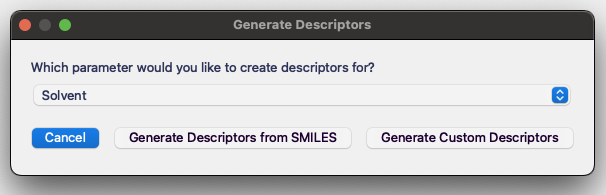
After selecting the parameter, you will need to copy in SMILES strings for all the possible compounds.
Note
SMILES (Simplified Molecular-Input Line-Entry System) are a great way to represent organic molecules using simple text.
Most chemical drawing software (ChemDraw, Marvin Sketch, Biovia Draw) has functionality to generate SMILES from chemical structure. You can also find SMILES in most online chemical databases, as well as in Wikipedia articles.
You can learn more about SMILES on Wikipedia.
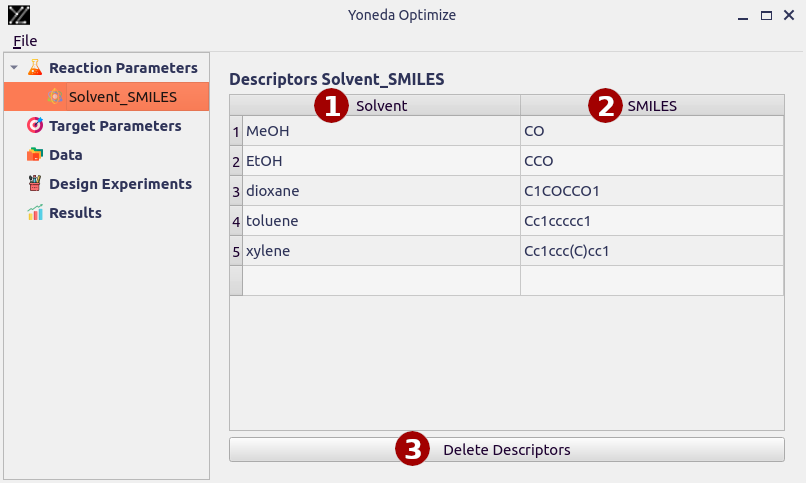
- Name of the parameter
This column contains names of allowed parameters.
If you edit, add or remove values, make sure the parameters are consistent with the
Valuescolumn on theReaction Parameterspage.
- SMILES
Paste the SMILES strings into this column. The SMILES strings will be used to generate chemical descriptors for the compounds.
- Delete Descriptors
This button deletes this table and associated descriptors.
Importing SMILES
If you already have SMILES strings stored in a .csv file, you can import them using the Import Descriptors button.
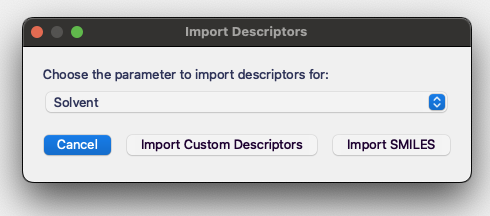
You will then need choose the Import SMILES option and upload the file.
- The SMILES need to be stored in a .csv file with two columns, with the following format:
entries are separated by commas
first line contains heading, the names of the columns are not used by Yoneda Optimize
first column contains names of the compounds, these need to match the
Valueson theReaction Parameterspagesecond column contains SMILES strings
Custom Chemical Descriptors
If you have specific insights into the reaction mechanism and how it’s affected by the properties of the compounds, you can design your own descriptors. However, this is an advanced feature as poorly chosen descriptors might hinder the quality of suggested experiments.
You can generate your own descriptors directly within Yoneda Optimize by clicking the Generate Descriptors button on the Reaction Parameters page. You will then need to choose the Generate Custom Descriptors option.

Afterward you will be able to generate your own descriptors. You should start by pressing the Add Column button to add a new property.
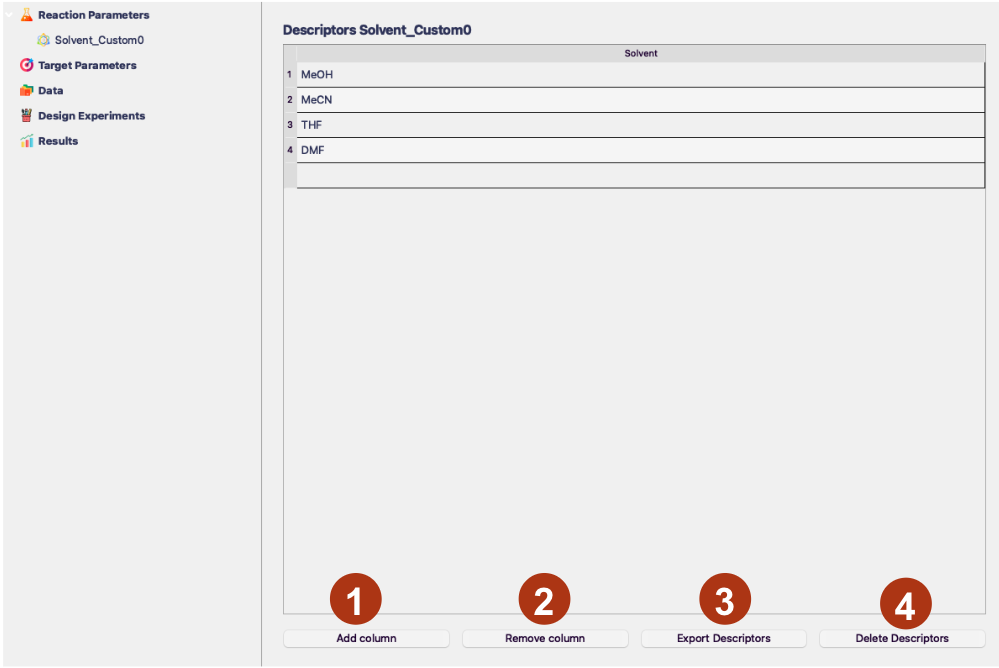
- Add column
This button adds a new column to the table representing the numerical properties you want associated with your values. You can add as many columns as you need.
- Remove column
This button allows you to remove the columns. You will have to specify which column you want to remove in a pop-up window.
- Export Descriptors
This button exports the descriptors to a .csv file, which you can then store locally and reuse between different projects.
- Delete Descriptors
This button deletes this table and associated descriptors.
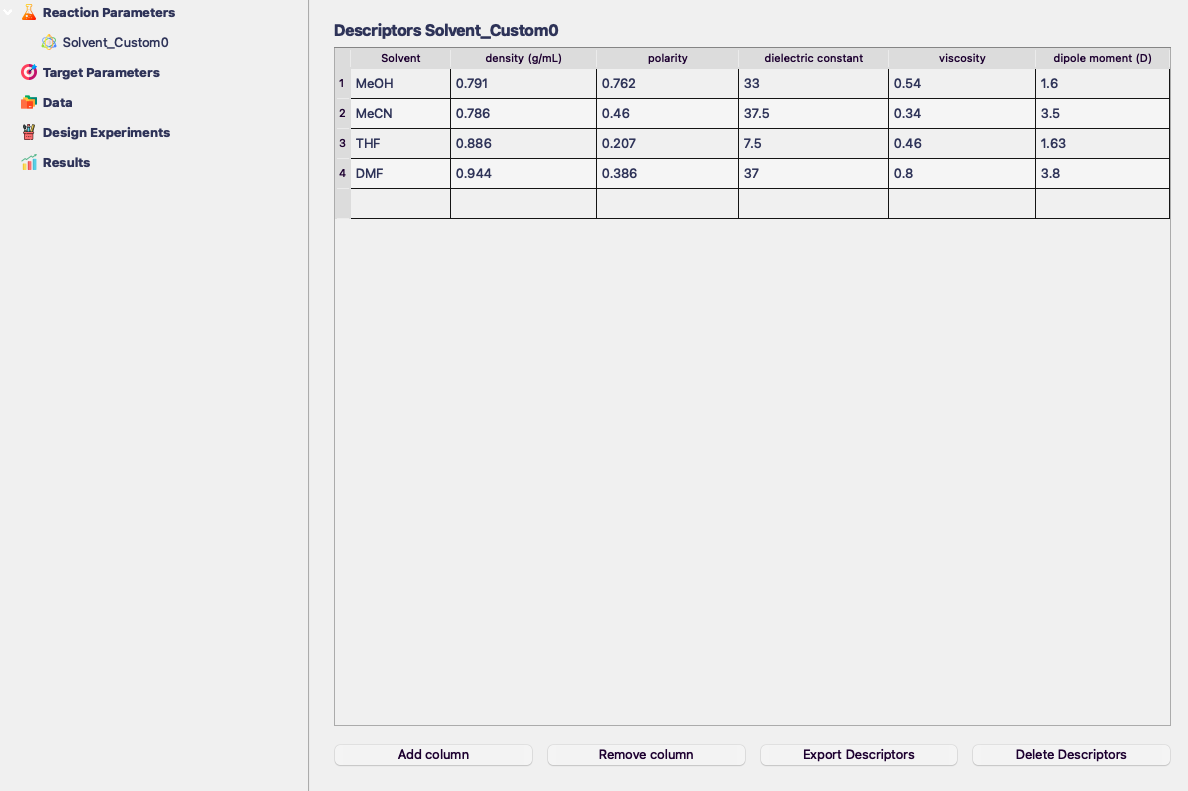
Importing Custom Descriptors
You can import your custom descriptors using the Import Descriptors button.
- The descriptors need to be stored in a .csv file with the following format:
entries are separated by commas
first line contains heading, the names of the columns are not used by Yoneda Optimize
first column contains names of the compounds, these need to match the
Valueson theReaction Parameterspagesubsequent columns contain descriptors, i.e. numeric entries